Cell Population Modeling of Oscillating Yeast Cultures
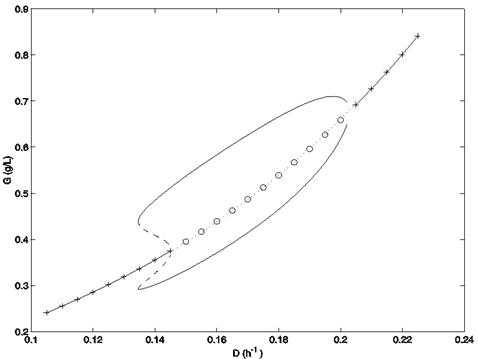
The objective of this project was the dynamic modeling of yeast cell populations which exhibit autonomous oscillations in continuous culture. The work focused on Saccharomyces cerevisiae due to the industrial significance of this microorganism and the availability of extensive data. We developed cell population balance equation (PBE) models of increasing sophistication to describe the synchronization of cell cycle dependent oscillations. Numerical bifurcation studies demonstrated that our most recent model is consistent with experiments in which a stable steady state and a stable limit cycle coexist over an observable range of dilution rates. Parameter estimation was pursued as a means to achieve quantitative agreement between the model and experimental data. Bifurcation behavior has been used to evaluate the accuracy of reduced-order dynamic models derived by applying proper orthogonal decomposition to spatiotemporal data generated by the cell population model.
More recently we developed a computationally tractable framework for incorporating intracellular structure within a cell population model. Rather than formulate the governing PBE, the cell population was described by a large ensemble of individual cell models which differ according to key properties such as the intracellular kinetics. The number distribution of any property captured by the individual cell model could be calculated by simulation of a sufficient number of individual cells. We used a 1000 cell ensemble to describe the synchronization and desynchronization of yeast glycolytic oscillations which result from various perturbations in the intracellular and extracellular environments. Our more recent work focused on the construction of much larger cell ensembles (~10000 cells) to evaluate a hypothesized synchronization mechanism for yeast metabolic oscillations involving the sulfate assimilation pathway.
Funding: National Science Foundation (BES-9522274, CTS-9501368), Alexander von Humboldt Foundation (Germany) and UMass
Students: Michael J. Kurtz (Ph.D.), Guang-Yan Zhu (Ph.D.) and Yongchun Zhang (Ph.D.)
Collaborators: Profs. Martin Hjortso (LSU), Matthias Reuss (Stuttgart) and Yannis Kevrekidis (Princeton)
Publications:
- Zamamiri, A. M., Y. Zhang, M. A. Henson and M. A. Hjortso, "Dynamic Analysis of an Age Distribution Model of Oscillating Yeast Cultures," Chemical Engineering Science, 57, 2169-2181 (2002). [PDF]
- Daoutidis, P. and M. A. Henson, "Dynamics and Control of Cell Populations in Continuous Bioreactors," AIChE Symposium Series, 326, 274-289 (2002). [PDF]
- Mhaskar, P., M. A. Henson and M. A. Hjortso, "Cell Population Modeling and Parameter Estimation of Continuous Cultures of Saccharomyces cerevisiae," Biotechnology Progress, 18, 1010-1026 (2002). [PDF]
- Henson, M. A., D. Muller and M. Reuss, "Cell Population Modeling of Yeast Glycolytic Oscillations," Biochemical Journal, 368, 433-446 (2002). [PDF]
- Zhang, Y., M. A. Henson and Y. Kevrekidis, "Nonlinear Model Reduction for Dynamic Analysis of Cell Population Models," Chemical Engineering Science, 58, 429-445 (2003). [PDF]
- Henson, M. A., "Dynamic Modeling and Control of Yeast Cell Populations in Continuous Biochemical Reactors," Computer and Chemical Engineering, 27, 1185-1199 (2003). [PDF]
- Henson, M. A., "Dynamic Modeling of Microbial Cell Populations," Current Opinion in Biotechnology, 14, 460-467 (2003). [PDF]
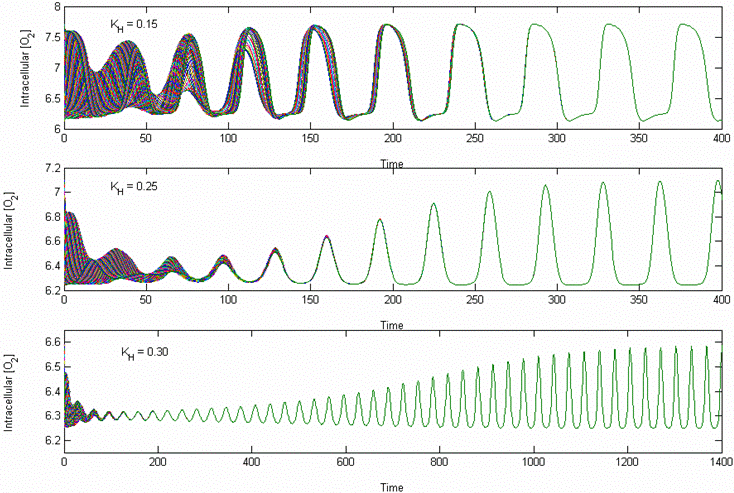
- Henson, M. A., "Modeling the Synchronization of Yeast Respiratory Oscillations," Journal of Theoretical Biology, 231, 443-458 (2004). [PDF]
- Henson, M. A., "Cell Ensemble Modeling of Metabolic Oscillations in Continuous Yeast Cultures," Computers and Chemical Engineering, 29, 645-661 (2005). [PDF]
- Bold K. A., Y. Zou, I. G. Kevrekidis and M. A. Henson, "Efficient Simulation of Coupled Biological Oscillators through Equation-Free Uncertainty Quantification" Journal of Mathematical Biology, 55, 331-352 (2007).[PDF]
Bifurcation Diagram for a Yeast Cell Cycle Oscillation Model
Cell Population Model Prediction of Yeast Respiratory Oscillations