Dynamics and Control of Biochemical Reactorst
We developed model-based techniques for dynamic analysis and feedback control of continuous biochemical reactors. Numerical bifurcation analysis was proposed as a tool for obtaining efficient and complete characterization of bioreactor model behavior. Application of this methodology to three previously published bioreactor models revealed unexpected steady-state and transient behaviors which can facilitate model development and validation. We also developed a nonlinear control strategy that allows stabilization of a desired steady state for mixed culture fermentations where two microbial populations compete for a common rate limiting substrate. The proposed method was successfully applied to a simulation model describing the production of two strains of Saccharomyces cerevisiae.
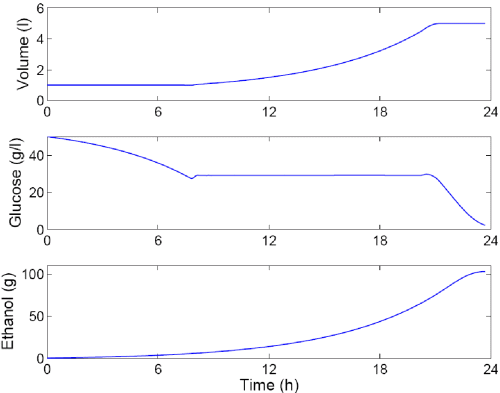
More recent work focused on the use of yeast cell population models for model-based optimization and control. A simple nonlinear control scheme based on a population balance equation model and feedback linearization was proposed for attenuation of sustained oscillations resulting from cell cycle synchronization. We have developed a more sophisticated model predictive control strategy that allows attenuation of oscillations that adversely affect bioreactor stability and/or induction of oscillations that lead to increased production of important metabolites synthesized only during part of the cell cycle. Cell-cycle dependent metabolite synthesis was further explored using a simple population balance model for dynamic optimization of ethanol production in fed-batch culture.
Funding: National Science Foundation (BES-9522274, CTS-9501368) and UMass
Students: Michael J. Kurtz (Ph.D.), Guang-Yan Zhu (Ph.D.), Yongchun Zhang (Ph.D.), Pavan Kumar Reddy Kambam (Ph.D.) and Jared Hjersted (5th year Ph.D.)
Collaborators: Martin Hjortso (LSU),Matthias Reuss (U. Stuttgart) and Lianhong Sun (University of Science and Technology of China)
Publications:
- Kurtz, M. J., M. A. Henson, and M. A. Hjortso, "Nonlinear Control of Competitive Mixed-Culture Bioreactors via Specific Cell Adhesion," Canadian Journal of Chemical Engineering, 78, 237-247 (2000). [PDF]
- Zhu, G.-Y., A. M. Zamamiri, M. A. Henson and M. A. Hjortso, "Model Predictive Control of Continuous Yeast Bioreactors Using Cell Population Models," Chemical Engineering Science, 55, 6155-6167 (2000). [PDF]
- Zhang, Y. and M. A. Henson, "Bifurcation Analysis of Continuous Biochemical Reactor Models," Biotechnology Progress, 17, 647-660 (2001). [PDF]
- Zhang, Y., A. M. Zamamiri, M. A. Henson and M. A. Hjortso, "Cell Population Models for Bifurcation Analysis and Nonlinear Control of Continuous Yeast Bioreactors," Journal of Process Control, 12, 721-734 (2002). [PDF]
- Henson, M. A., "Distribution Control of Particulate Systems Based on Population Balance Equation Models," Proc. American Control Conference, Denver, CO, 3967-3972 (2003). [PDF]
- Henson, M. A., D. Muller and M. Reuss, "Combined Metabolic and Cell Population Modeling for Yeast Bioreactor Control," Proc. IFAC Symposium on Advanced Control of Chemical Processes, Hong Kong (2004). [PDF]
- Hjersted, J. and M. A. Henson, "Population Modeling for Ethanol Productivity Optimization in Fed-Batch Yeast Fermenters," Proc. American Control Conference, Portland, OR (2005).
- Henson, M. A., "Biochemical Reactor Modeling and Control: Exploiting Cellular Biology to Manufacture High Value Products," IEEE Control Systems Magazine, 26, 54-62, August (2006). [PDF]
- Kambam, P. K. R., M. A. Henson and L. Sun, "Design and Mathematical Modeling of a Synthetic Symbiotic Ecosystem," IET Systems Biology, 2, 33-38 (2008). [PDF]
- Henson, M. A., "Model-Based Control of Biochemical Reactors," in The Control Handbook, 2nd edition, William Levine (Ed.), Taylor and Francis, New York, NY, 2010.
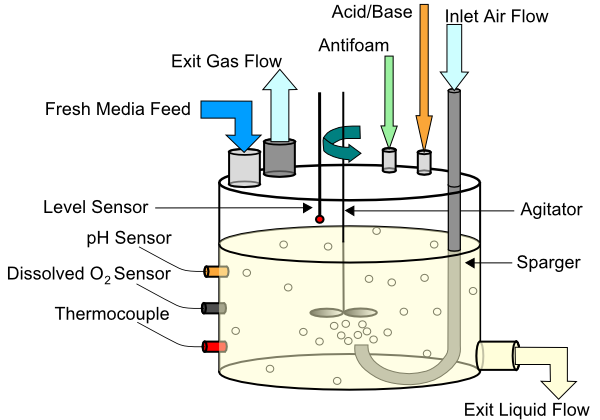
Continuous Biochemical Reactor
Optimal Substrate Feeding Policy for Cell Cycle Dependent Ethanol Production