Biological Timekeeping
The circadian clock generates 24 hour rhythms that drive physiological and behavioral processes in a diverse range of organisms including microbes, plants, insects and mammals. Circadian rhythms may be disrupted in otherwise healthy individuals by a variety of perturbations including jet lag, social jet lag, rotating shift work, seasonal changes and altered brain function. Circadian disruptions have been associated with sleep loss, erratic wake times, reduced cognitive ability, obesity, diabetes, heart attacks, schizophrenia and Alzheimer’s disease. In mammals, circadian rhythms are generated by an internal self-sustained oscillator located in the hypothalamic suprachiasmatic nucleus (SCN) and consisting of approximately 20,000 heterogeneous neurons. Recent experimental advances have produced improved understanding of the molecular mechanisms involved in circadian rhythm generation at the single cell level. However, the intercellular mechanisms that allow large populations of coupled pacemaker cells to synchronize and coordinate their rhythms remain poorly understood.
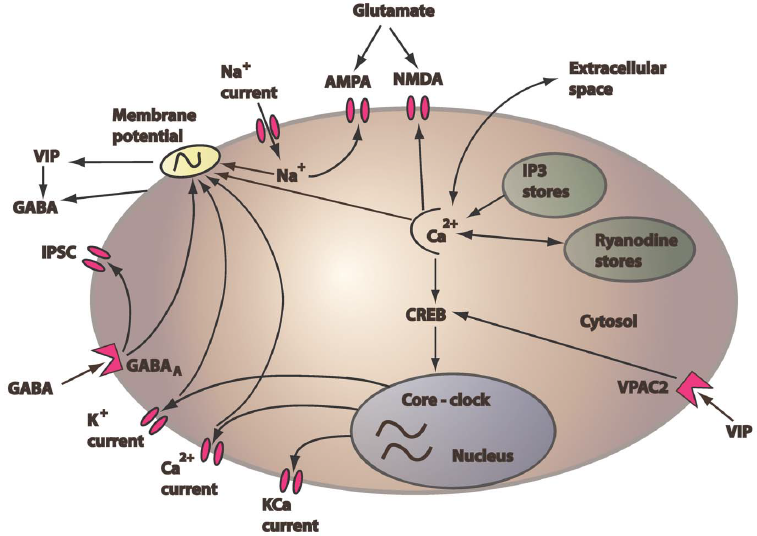
We are developing multicellular network models to better understand neurotransmitter mediated intracellular communication within the SCN. Our single neuron model accounts both for the gene regulatory circuit that generates autonomous oscillations and the electrophysiology that produces neural firing. Multicellular models are developed by coupling a heterogeneous collection of individual neurons according to a prescribed network connectivity pattern, which is derived from experiment to the extent possible. The resulting models are characterized by a high degree of heterogeneity with respect to single cell periodicity, neurotransmitter release and spatial organization, mimicking structural patterns within the SCN associated with distinct circadian functions. Dynamic simulations are performed to better understand the role of cellular and network heterogeneities on intercellular synchronization and entrainment to environmental cues.
Project funding is generously provided by:
Current projects include:
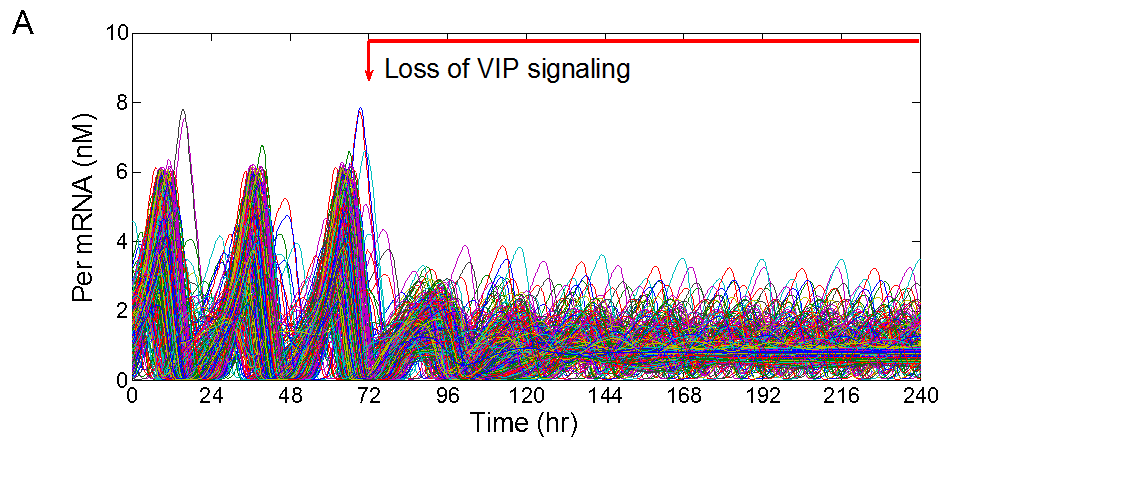
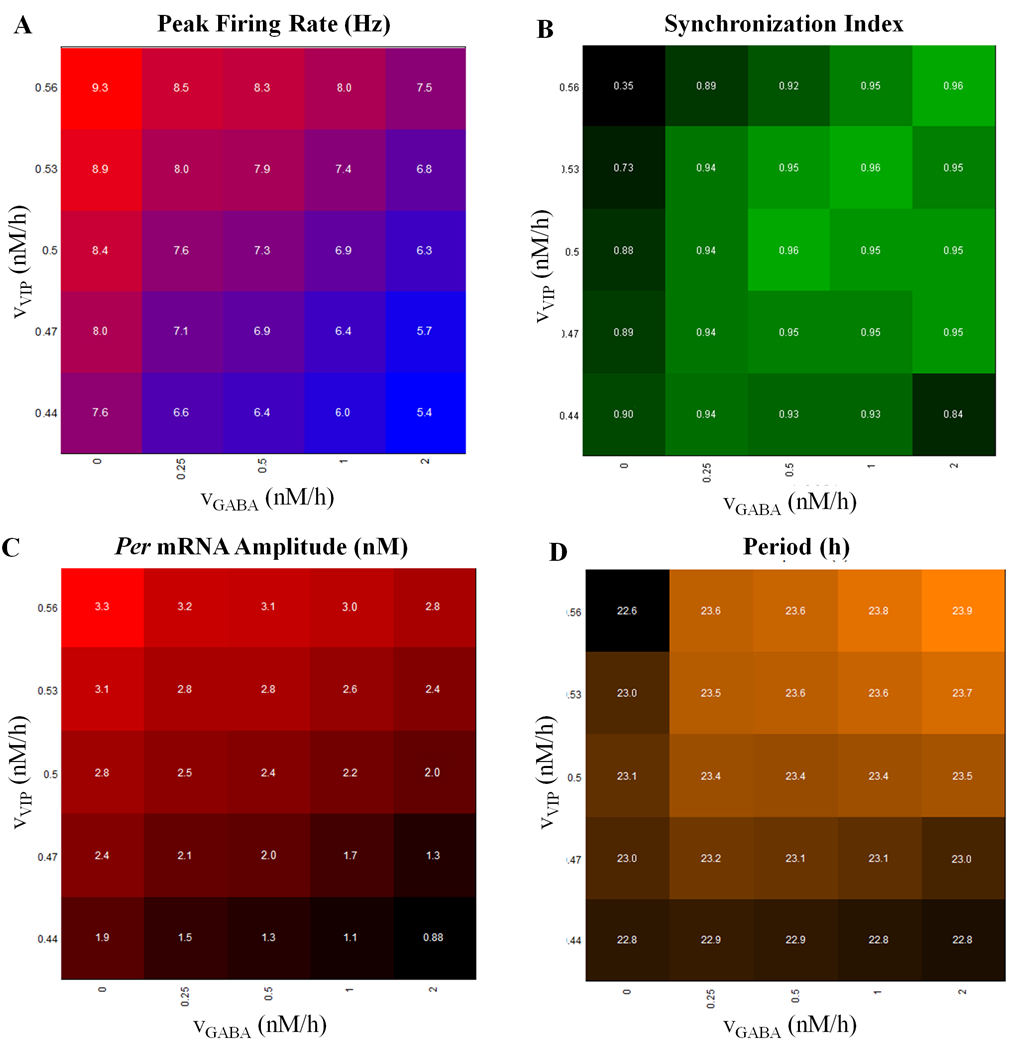
- Development of multicellular models that predict the differential roles of the neurotransmitters vasoactive intestinal polypeptide and γ-aminobutyric acid in intercellular synchronization and light entrainment.
Collaborator: Erik Herzog, Department of Biology, Washington University
- Development and testing of equation-free simulation methods to dramatically reduce computation times such that large neural networks and many types of heterogeneity can be efficiently explored.
Collaborator: Yannis Kevrekidis, Department of Chemical and Biological Engineering, Princeton University
Selected publications:
- Vasalou C. and M. A. Henson, "A Multiscale Model to Investigate Circadian Rhythmicity of Pacemaker Neurons in the Suprachiasmatic Nucleus,'' PLOS Computational Biology, 6: e1000706. doi:10.1371/journal.pcbi.1000706 (2010). [PDF]
- Vasalou, C., E. D. Herzog and M. A. Henson, "A Model for Intercellular Synchronization in Circadian Neural Networks," Biophysical Journal, 101, 12-20 (2011). [PDF]
- Vasalou, C. and M. A. Henson, “A Multicellular Model for Differential Regulation of Circadian Signals in the Shell and Core Regions of the SCN,” Journal of Theoretical Biology, 288, 44-56 (2011). [PDF]
- Henson, M. A., “Multicellular Models of Intercellular Synchronization in Circadian Neural Networks,” Chaos, Solitons and Fractals, 50, 48-64 (2013). [PDF]
- Su, J. and M. A. Henson, Circadian Gating of the Mammalian Cell Cycle Restriction Point: A Mathematical Analysis,” IEEE Life Science Letters, 1, 11-14, doi:10.1109/LLS.2015.2449511 (2015) [Link]
- Kingsbury, N. J., S. R. Taylor and M. A. Henson, “Inhibitory and Excitatory Networks Balance Cell Coupling in the Suprachiasmatic Nucleus: A Modeling Approach,” Journal of Theoretical Biology, 397, 135-44, doi: 10.1016/j.jtbi.2016.02.039 (2016). [Link]