Renewable Biofuels and Biochemicals
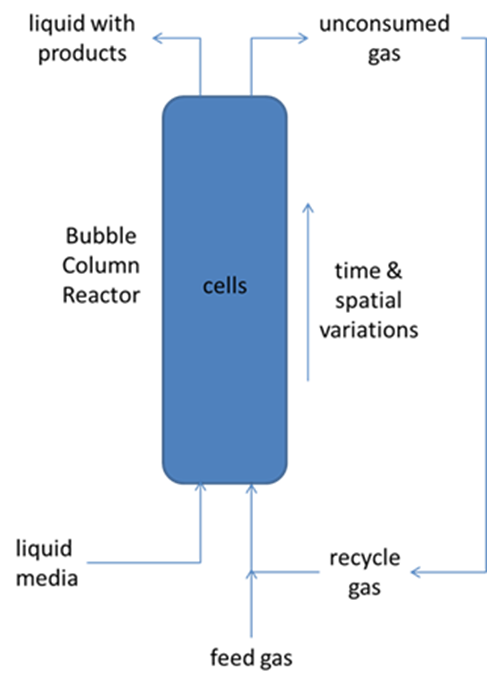
An essential component of the quest for energy independence is to develop renewable, environmentally friendly sources of biofuels and biochemicals via the conversion of abundant plant biomass and waste streams. An emerging conversion route with wide feedstock versatility is direct fermentation of waste gas streams and synthesis gas (syngas; mainly comprised of H2/CO/CO2) by specialized CO fermenting microbes. Because syngas can be produced relatively cheaply from a wide variety of biomass feedstocks, the bottleneck in this route is the syngas fermentation step. Key considerations include the metabolic capabilities of the microbial catalyst that converts syngas into the desired biochemical, gas-liquid mass transfer characteristics that determine the availability of soluble gas components for microbial conversion and the bioreactor design that affects all aspects of the conversion process. Commercial development favors bubble column reactors due to their high average mass transfer driving forces and long gas-liquid contact times.
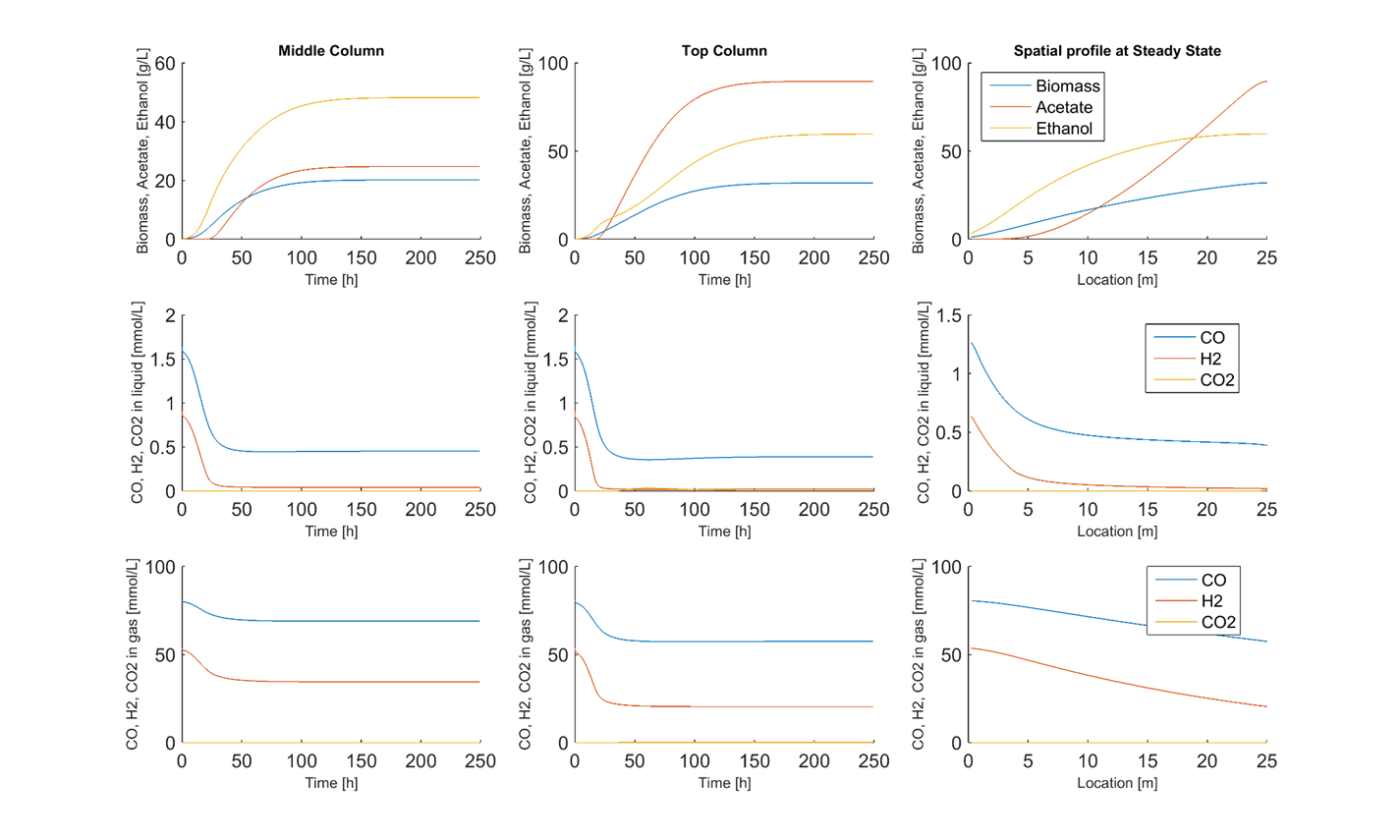
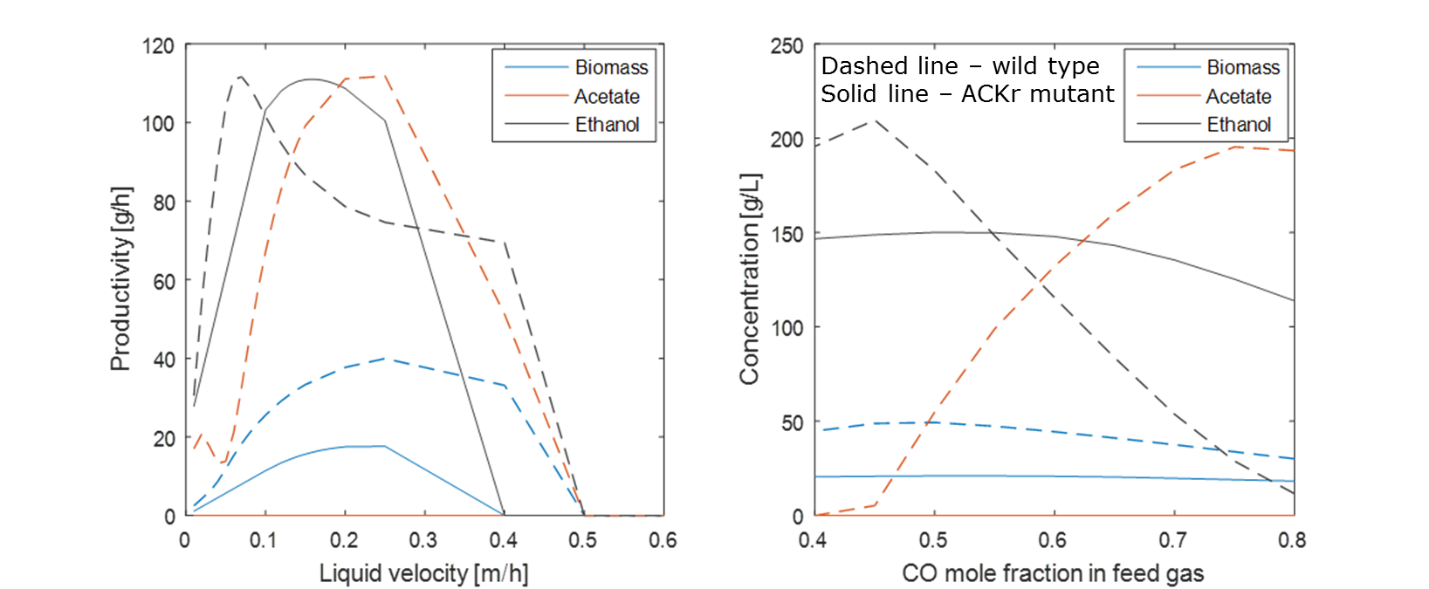
We are utilizing genome-scale metabolic reconstructions of CO fermenting bacteria to develop combined metabolic-transport models that capture the complex spatiotemporal behavior of syngas bubble column reactors. The bioreactor models are formulated by combining genome-scale reconstructions with transport equations that capture the key hydrodynamics of the multiphase gas-liquid system. Our current models provide both spatial and temporal predictions of cellular growth, dissolved gas consumption, byproduct synthesis, gas-liquid mass transfer and gas phase hydrodynamics. The models consisting of partial differential equations with embedded linear programs are solved by discretizing the spatial domain and integrating the time-dependent model using DFBAlab within the MATLAB environment. Simulations are performed to analyze the effects of column operating conditions and microbial engineering strategies on growth rates, product titers and volumetric productivities.
Project funding is generously provided by:
Current projects include:
- Development and experimental testing of bubble column reactor models with the CO fermenting bacteria Clostridium ljungdahlii and Clostridium autoethanogenum.
Collaborator: Derek Griffin and Bruce Li, Lanzatech.
- Integration of microbial and catalytic technologies for development of hybrid processes for biomass conversion to fuels and chemicals.
Collaborator: Friederike Jentoft, Department of Chemical Engineering, UMass and Emma Wylde,, European Bioenergy Research Institute, Aston University.
Selected publications:
- Kambam, P. K. R. and M. A. Henson, "Engineering Bacterial Processes for Cellulosic Ethanol Production," Biofuels, 1, 729-744 (2010). [Link]
- Hanly, T. J. and M. A. Henson, "Dynamic Flux Balance Modeling of Microbial Co-Cultures for Efficient Batch Fermentation of Glucose and Xylose Mixtures," Biotechnology and Bioengineering, 108, 376-385 (2010). [PDF]
- Hanly, T. J., M. Urello and M. A. Henson, “Dynamic Flux Balance Modeling of S. cerevisiae and E. coli Co-cultures for Efficient Consumption of Glucose/Xylose Mixtures,” Applied Microbiology and Biotechnology, 93, 2529-2541 (2012). [Link]
- Mahadevan, R. and M. A. Henson, “Genome-based Modeling and Design of Metabolic Interactions in Microbial Communities,” Computational and Structural Biotechnology Journal, 3: e201210008. dx.doi.org/10.5936/csbj.201210008 (2012). [PDF]
- Hanly, T. J. and M. A. Henson, “Dynamic Metabolic Modeling of a Microaerobic Yeast Co-culture: Predicting and Optimizing Ethanol Production from Glucose/Xylose Mixtures,” Biotechnology for Biofuels, 6, 44, doi:10.1186/1754-6834-6-44 (2013). [Link]
- Henson Chen, J., J. A. Gomez, K. Hoffner, P. I. Barton and M. A. Henson, “Metabolic Modeling of Synthesis Gas Fermentation in Bubble Column Reactors,” Biotechnology for Biofuels, 8, 89, doi:10.1186/s13068-015-0272-5 (2015). [Link]
- Henson, M. A., “Genome-Scale Modeling of Microbial Metabolism with Temporal and Spatial Resolution,” Biochemical Society Transactions, 43, 1164-1171, doi:10.1042/BST20150146 (2015). [Link]
- Chen, J., P. Phalak, J. A. Gomez, K. Hoffner, P. I. Barton and M. A. Henson, “Spatiotemporal Modeling of Microbial Metabolism,'' BMC Systems Biology, 10:21, doi: 10.1186/s12918-016-0259-2 (2016). [PDF]
- Chen, J. and M. A. Henson, In silico Metabolic Engineering of Clostridium ljungdahlii for Synthesis Gas Fermentation,” [Submitted]