Microbial Communities and Biofilms
Microbial biofilms are critically important in medical, environmental and engineered biological systems. For example, the human gut microbiome has emerged as a major focus for biomedical research with mounting evidence suggesting unhealthy gut flora biofilms are associated with illnesses including autoimmune diseases, colorectal cancer and inflammatory bowel disease. Environmental microbial biofilms form the basis of global nutrient cycles from nitrogen fixation to carbon fluxes. Additionally, the study of natural biofilms has gained in popularity recently due to their efficient organization and ability to optimize multiple tasks simultaneously like the deconstruction of complex, recalcitrant plant materials into simple sugars. A major goal of current biofuels research is to engineer microbes that mimic these naturally occurring biofilms for biomass conversion to renewable liquid fuels. While foundational to the vast majority of microbial life on the planet, the basic design principles of microbial biofilms are still poorly understood.
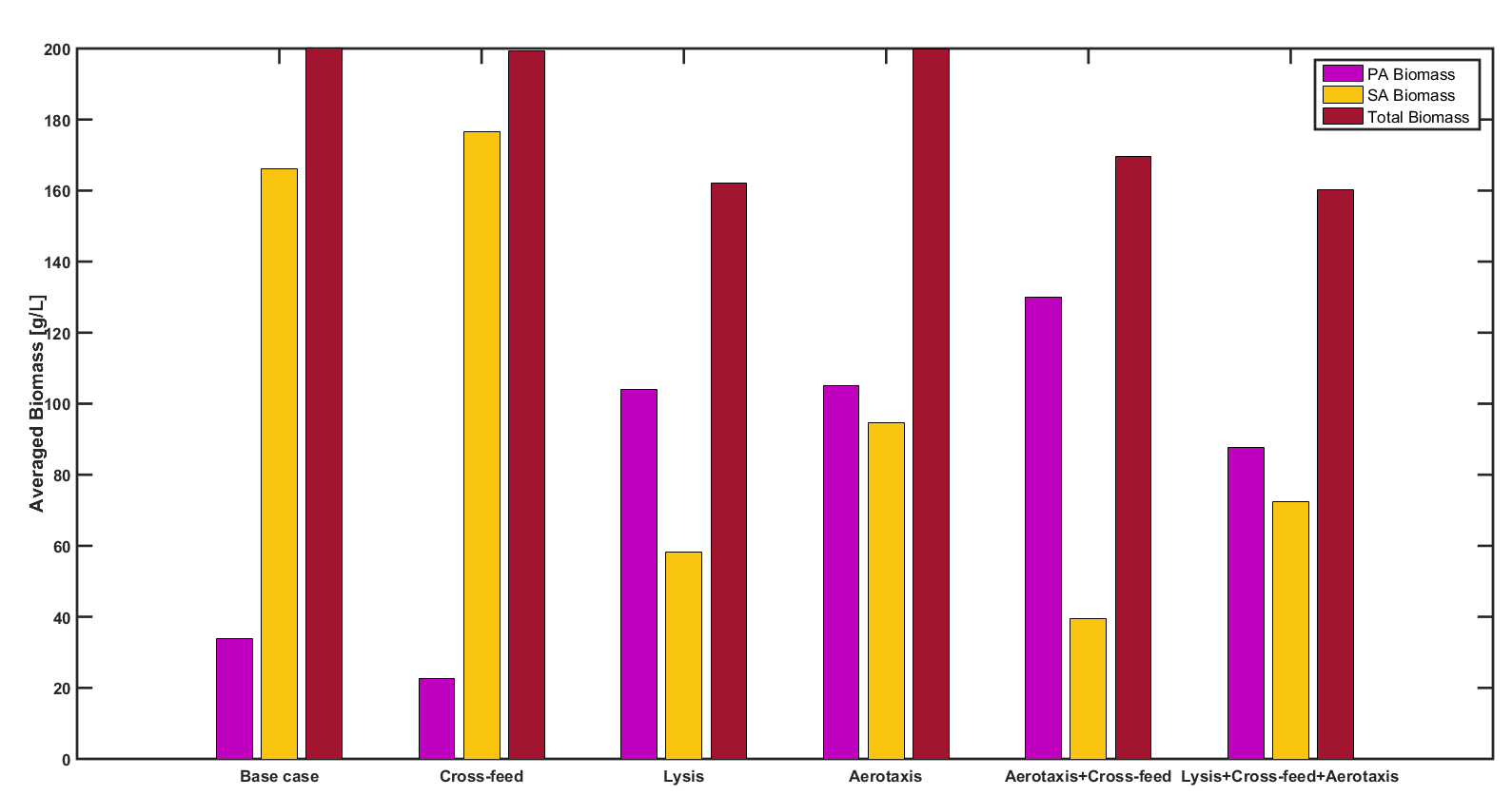
We are developing metabolic models of single species and multispecies biofilms that predict temporal and spatial variations with genome-scale resolution. The biofilm models are formulated by combining genome-scale reconstructions of species metabolism with transport equations that capture the relevant diffusional and convective processes within the biofilm. Our current models capture cellular growth and death, biofilm expansion and contraction, competition for nutrients, cross-feeding of metabolic byproducts, chemotaxis of motile species, secretion of small molecule inhibitors and antibiotic treatment. The biofilm models consisting of partial differential equations with embedded linear programs are solved by discretizing the spatial domain and integrating the time-dependent model using DFBAlab within the MATLAB environment. Simulations are performed to analyze the understand biofilm design principles and to develop improved antibiotic treatment strategies for biofilm eradication.
Funding:
Current projects include:
- Clostridium difficile biofilms implicated in recurring infections in the gastrointestinal tracts of patients previously treated with oral antibiotics that disrupt the healthy gut microbiome.
Collaborator: Adam Driks, Department of Microbiology and Immunology, Loyola University Chicago.
- Pseudomonas aeruginosa and multispecies biofilms responsible for chronic lung infections in cystic fibrosis patients.
Collaborator: George O’Toole, Department of Microbiology and Immunology, Dartmouth University.
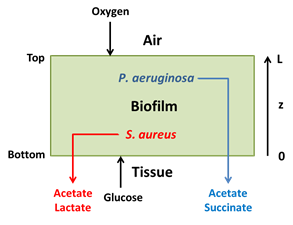
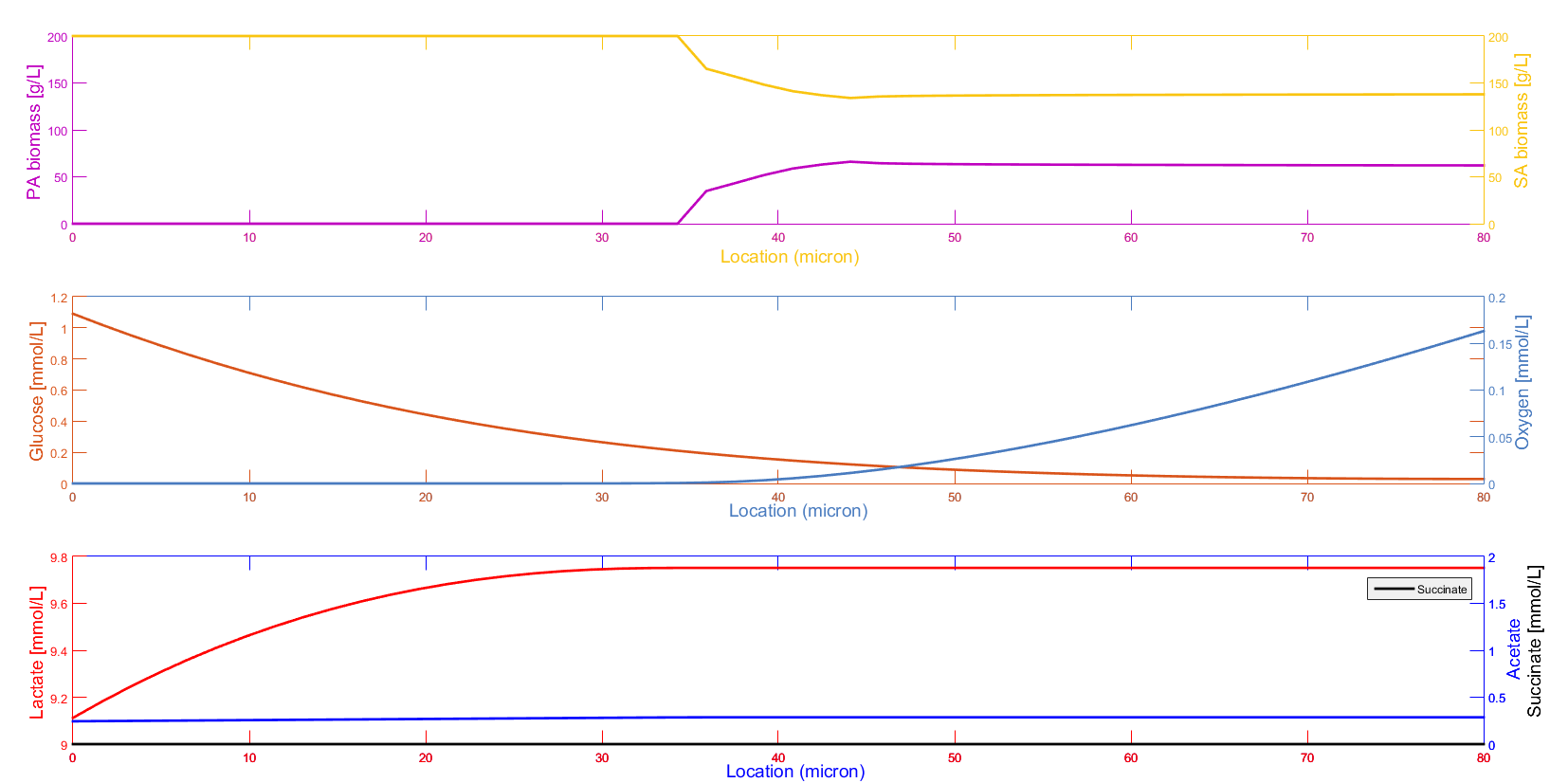
- Pseudomonas aeruginosa/Staphylococcus aureus and other multispecies biofilms responsible for the chronic wound infections in patients with diabetes and other circulatory problems.
Collaborator: Ross Carlson, Center for Biofilm Engineering, Montana State University
- Engineered multispecies biofilms which robustly convert plant derived cellulose into the biofuel isobutanol despite environmental perturbations and the presence of competing species.
Collaborator: Ross Carlson, Center for Biofilm Engineering, Montana State University
Selected publications:
- Henson, M. A., “Genome-Scale Modeling of Microbial Metabolism with Temporal and Spatial Resolution,” Biochemical Society Transactions, 43, 1164-1171, doi:10.1042/BST20150146 (2015). [Link]
- Chen, J., P. Phalak, J. A. Gomez, K. Hoffner, P. I. Barton and M. A. Henson, “Spatiotemporal Modeling of Microbial Metabolism,'' BMC Systems Biology, 10:21, doi: 10.1186/s12918-016-0259-2 (2016). [Link]
- Phalak, P., J. Chen, R. P. Carlson and M. A. Henson, “Spatiotemporal Metabolic Modeling of a Chronic Wound Biofilm Consortium”. [Submitted]
- Henson, M. A. and P. Phalak, In silico Analysis of Clostridium difficile Biofilm Metabolism and Antibiotic Treatment. [Submitted]
- Phalak, P., J. Chen, R. P. Carlson and M. A. Henson, “Metabolic Modeling of a Chronic Wound Biofilm Consortium Predicts Spatial Partitioning of Bacterial Species'' . [PDF]